
|
Published: 14 May 2012
Slowing the development of antibiotic resistance
New research on how proteins are exported by disease-causing bacteria may pave the way for new antibiotics that disarm bacteria rather than killing them, a potentially important step in the fight against antibiotic resistance.
|
|
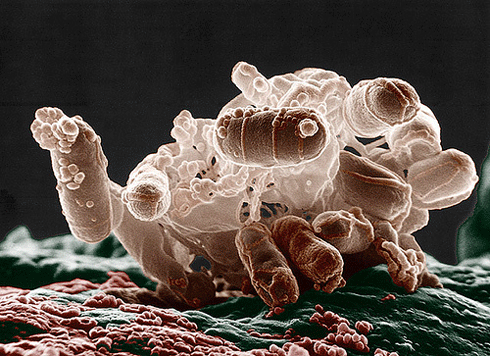
A cluster of Escherichia coli bacteria under an electron microscope. New research into how disease-causing bacteria such as E. coli export particular proteins may lead to new drugs that will not promote antibiotic resistance at the same rate as the current generation of antibiotics. Credit:
Microbeworld Rights: Licensed under a Creative Commons Attribution Non-Commercial Share-Alike License |
One of the ways that disease causing bacteria infect hosts is by exporting proteins. New research, published in Nature Structural and Molecular Biology, has discovered a ‘translocation and assembly module’ (TAM) that allows bacteria to shuttle key disease-promoting molecules from inside the bacterial cell, where they are made, to the outside surface. There these molecules prime the bacteria for infection by helping the bacteria adhere to our bodies and by performing other disease-related functions.
‘The TAM was discovered in many disease-causing bacteria, from micro-organisms that cause whooping cough and meningitis, to hospital-acquired bacteria that are developing resistance to current antibiotics,’ says lead author and PhD student Joel Selkrig.
The international team behind the study showed that the TAM was made of two protein parts, TamA and TamB, which function together to form a machine of molecular scale.
Normal virulent bacteria were then compared to mutant strains of bacteria engineered to have no TAM.
‘We noticed that proteins important for disease were missing in the outer membrane of the mutant bacteria,’ Mr Selkrig says.
Mr Selkrig says the next step for the group is to dissect the molecular mechanism of how the TAM complex functions and then, in collaboration with researchers at the Monash Institute of Pharmaceutical Sciences, design an antibiotic that inhibits the TAM in bacteria.
Antiobiotic resistance develops through a process of natural selection. Mutant bacteria resistant to antibiotics exist naturally in bacterial populations, but in low numbers. When bacterial populations are subjected to drugs that cause high mortality, those bacteria that survive to reproduce are the resistant mutant strains, which then multiply rapidly in the absence of control.
‘A drug designed to inhibit TAM function would unlikely kill bacteria, but simply deprive them of their molecular weaponry, and in doing so, disable the disease process. By allowing bacteria to stay alive after antibiotic treatment, we believe we can also prevent the emergence of antibiotic resistance, which is fast becoming a major problem worldwide,’ says Mr Selkrig.
Source: Monash University



